Constructing Biological Motor Powered Nanomechanical Devices
by
Carlo Montemagno*, George Bachand, Scott Stelick, Marlene Bachand
Department of Agricultural and Biological Engineering
Cornell University, Ithaca, NY 14853
*Corresponding author: [email protected]
Web page: http://falcon.aben.cornell.edu/
This is a draft paper for the
Sixth
Foresight Conference on Molecular Nanotechnology.
The final version was published in the special Conference issue of Nanotechnology:
Nanotechnology 10 (1999) 225-231.
Abstract
A confluence in scientific advancements associated with molecular biology and nanofabrication technology now offer, for the first time, the potential of engineering functional hybrid organic/inorganic nanomechanical systems. Our long-term goal is the integration of the biological motor, F1-ATPase with nano-electro-mechanical systems (NEMS) to create a new class of hybrid nanomechanical devices. The F1-ATPase biomolecular motor is capable of generating a force >100 pN, has a calculated no-load rotational velocity of 17 r.p.s., and a diameter of less than 12 nm. These characteristics are consistent with the engineering features associated with currently producable nanomechanical structures. Thus, the potential to seamlessly integrate the motive power of life with engineered nanofabricated devices now exists. This paper will address current research on the integration of the F1-ATPase motor protein with NEMS specifically designed to evaluate motor performance.
Introduction
Scientific advancements in both molecular biology and nanofabrication technology now provide the potential of engineering functional hybrid organic/inorganic nanomechanical systems. Scientists have been studying a wide range of organic molecular motors for some time. Concurrently, inorganic, primarily silicon based, micromechanical devices have been pursued as useful devices. Only very recently has the size scale of nanofabricated inorganic mechanical devices approached a size scale that could conceivably be compatible with the force production and dimensions of molecular motors. Our long-term objective is to utilize the best attributes associated with both the organic and inorganic worlds for the examination and creation of nano-electro-mechanical systems (NEMS) that are powered by biological motors and chemical energy sources. We envision that F1-ATPase motors will pump fluids, and open and close valves in microfluidic devices, and provide mechanical drives for a new class of nanomechanical devices.
The rotary motion of the g subunit of F1-ATPase in response to the synthesis/hydrolysis of ATP has been previously demonstrated (Noji et al., 1997). The force generated by this motor protein was >100 pN, which is among the greatest of any known molecular motor. With a calculated no-load rotational velocity of 17 r.p.s. and a diameter of less than 12 nm (Sabbert et al., 1996; Yashuda et al., 1997), the F1-ATPase protein is a tailor made nano-motor. These properties, coupled with the fact F1-ATPase is automatically synthesized using the machinery of life, opens the door to the potential for creating chemically powered nanomechanical devices. Integration of nanoscale biocompatible lithographic processes with biological molecular motors may provide the means for creating a transparent interface between the organic/inorganic world. Insertion and self-assembly of hybrid ATPase-powered NEMS in host cells may be possible by taking advantage of cell physiological processes. In addition, host cells may provide power for the device in the form of ATP, as well as maintain a system for replacing the molecular motors when function ceases.
ATPase is a ubiquitous enzyme that is found in virtually every living organism. It consists of two separate portions: (1) F0, the hydrophobic, membrane-bound portion that is responsible for proton translocation, and (2) F1, the hydrophilic portion that is responsible for ATP synthesis and hydrolysis. As protons flow through the F0, the g subunit of the F1-ATPase rotates clockwise and ATP is synthesized. Hydrolysis of ATP results in counterclockwise rotation of the g subunit, and drives the reverse flow of protons. The a, b, and c subunits of the F0-ATPase form the channel which allows protons to flow through the membrane. The nucleotide binding and catalytic sites are located on the three a and three b subunits of the F1-ATPase, respectively (Kinosita et al., 1998). The g subunit is centrally located within the a3b3 hexamer, and rotates as a function of ATP synthesis/hydrolysis.
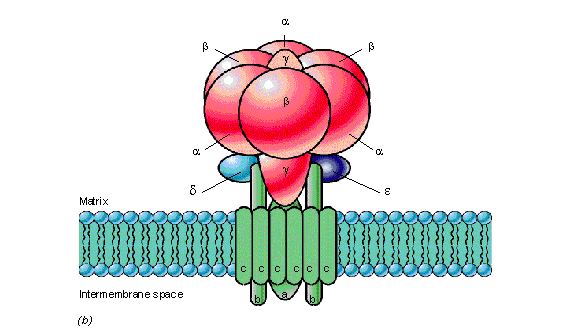
During hydrolysis, counterclockwise rotation of the g subunit provides interaction with all three forms of the b subunits in the order: AMP-PNP > ADP > empty form. The exact mechanism of interaction has yet to be determined. Crystallization of of the F1-ATPase has revealed that all three sites must contain bound nucleotides in order for rotation of the g subunit to occur (Bianchet et al., 1998). Further, the g subunit is displaced from its central axis during rotation.
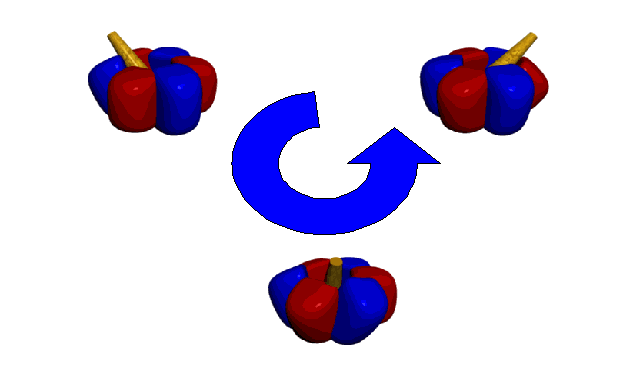
Despite the superb performance of the F1-ATPase motor protein, little is truly known about how this enzyme generates rotary motion. Neither the useful life of the motor nor the impact of local environmental variables such as pH and temperature on enzyme activity has been determined. The impact of motor generated waste products (i.e., protons and heat), as well as the effects of load on the performance and life of the motor need to be identified. A rigorous evaluation of the engineering properties of the F1-ATPase motor protein necessitates the development of assays that will provide consistent measurements of the performance of the F1-ATPase motor protein under different operating conditions. Our current research effort has focused on integrating the F1-ATPase with NEMS specifically designed to evaluate motor performance. We will present the results of this effort to construct a hybrid organic/inorganic nanoscale system that both provides insight into the basic mechanics of motor protein motion and establishes a technological foundation for functionally integrating these molecules with manufactured devices.
Integration of biological motors and NEMS
Platforms for the production of both biomolecular motors and
NEMS must be established in order to integrate these technologies
and produce hybrid systems. Initial research efforts, therefore,
have focused on the development of these platforms. In addition,
we also have begun evaluating the engineering properties of F1-ATPase.
Cloning and Expression of F1-ATPase from Bacillus PS3
The F1-ATPase coding sequence
(a, b, and g
subunits) was isolated from the thermophilic bacterium, Bacillus
PS3 (Matsui and Yoshida 1995), using the polymerase chain reaction
(PCR). Restriction endonucleases sites, BamHI and PstI,
were added to the 5' and 3' end of the coding sequence, respectively,
and used to directionally clone the 3.9 kb PCR product into the
plasmid pGEM-3Z f(-). Subsequently, site directed mutagenesis
was used to: (1) change the aCys193
to Ser, (2) change the gSer107 to Cys,
(3) change the g initiation codon from
GTG to ATG, and (4) change the g termination
codon from TAG to TAA. In addition, a ten histidine (His) tag
was inserted immediately downstream of the b
initiation codon. The mutated construct, pGEM-AITG, was then cloned
into the expression plasmid pQE-30, which added a six His tag
to the N-terminus of the a subunit.
The expression plasmid pQE-MH was inserted into Escherichia
coli JM103 D(uncB-uncD)
in which the majority of F1-ATPase
genes have been eliminated; thus, minimizing/eliminating the formation
of chimeric F1-ATPases.
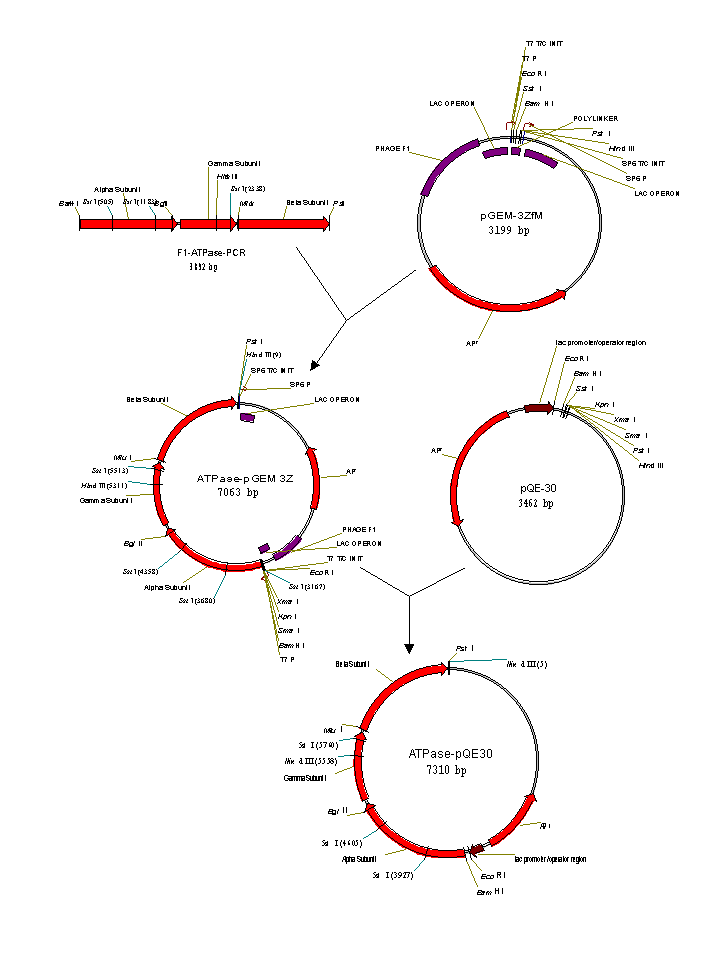
Previous research demonstrated a 40% reduction in enzymatic
activity associated with the addition of His tags to the F1-ATPase from E. coli (Ekuni
et al. 1998). For this reason, the mutated F1-ATPase
construct without His tags on the b
subunit also was cloned into pQE-30, and expressed in E. coli
JM103. Further, the His tags on the a
subunit can be removed from the recombinant F1-ATPase
using thrombin, which cleaves directly downstream of the His tags.
This expression system (pQE-M) will be used to evaluate the effect
of the His tags on both the a and b subunits on motor performance. Together,
pQE-M and pQE-MH provide flexibility in assessing motor performance
and the effects of adding attachment handles to particular regions
of the enzyme.
Expression of the recombinant F1-ATPase
was induced by the addition of 1 mM IPTG approximately 3 hours
after inoculation of M9 minimal media. Native protein was extracted
using lysozyme/sonication, and purified using Ni2+-NTA
affinity chromatography. Approximately 50 mg of F1-ATPase
was purified per liter of cell extract, and analyzed using SDS-polyacrylamide
gel electrophoresis. The activity of the purified protein was
measured using an ATP regeneration assay (Matsui and Yoshida,
1995; Matsui et al., 1997).

Attachment of biological molecules to nanofabricated
substrates
In order to integrate biomolecular motors into NEMS, procedures
for the specific attachment and positioning of these motors is
essential. Therefore, the objective of this experiment was to
evaluate the binding of biological molecules to nanofabricated
substrates. Electron beam lithography was utilized to etch an
array pattern on a 25 mm coverslip that had been coated with a
resist bilayer. Coverslips then were patterned with metal substrates
using evaporative deposition of gold, copper, or nickel. Subsequently,
the bilayer was removed to expose the array.

A six His-tag peptide was covalently coupled to carboxylate-modified
1 mm fluorescent microspheres using a water-soluble carbodiimide.
The His-tagged microspheres were allowed to attach to gold-, copper-,
and nickel-coated coverslips for 15 minutes at room temperature.
Unattached microspheres were removed through a series of washes,
and coverslips were observed using fluorescence microscopy. His-tagged
microspheres attached to all three substrates; however, attachment
was most frequently observed with nickel-coated coverslips.
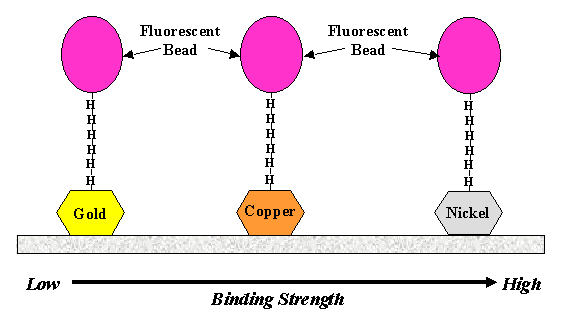
To test the strength of attachment, laser tweezers were used
to remove the microspheres from the substrate. The laser tweezers,
however, were unable to remove microspheres from any of the three
substrates suggesting that the bonding strength was greater than
600 pN. Further attempts to remove the microspheres with high
velocity flow suggest that the bonding strength increases from
gold to unoxidized copper to nickel. Oxidized copper does not
serve as a suitable surface for binding of His-tagged microspheres.
These experiments demonstrate a chemical mechanism for protein
binding and positioning to engineered structures that are compatible
with current nanofabrication technologies. Using this knowledge
in conjunction with standard e-beam lithographic methods (Craighead
and Mankiewich, 1982; Lercel et al., 1995; Lercel et al., 1996;
Carr and Craighead, 1997) we can now attach individual motor protein
molecules with a precision greater than 30 nm.
Attachment and movement of individual biomolecular
motors
Although the biological and chemical aspects of F1-ATPase have been studied, relatively little is know about the engineering properties of this enzyme. The objectives of this experiment were to: (i) attach F1-ATPase to a nanofabricated substrate, and (ii) measure the rotational velocity and angle of deformation of the g subunit. Analysis of crystallized F1-ATPase suggests that the g subunit is displaced from the central axis during rotation a distance >20 Å (Bianchet et al., 1998). By attaching a 1 mm microsphere to the g subunit, the displacement and angle of deformation of the g subunit can be determined by measuring the radial displacement of the microsphere. The angle of deformation will provide valuable insight on the mechanism behind rotation of the g subunit.

The g subunit of the recombinant F1-ATPase was specifically biotinylated through disulfide linkage to the gCys. The biotinylated protein then was attached to an array of 30 nm gold dots deposited on a coverslip. Fluorescent 1 mm microspheres coated with streptavidin were allowed to bind to the biotinylated g subunits. Subsequently, unattached microspheres were removed through a series of washes. Rotation of the g subunit was initiated by the addition of 2 mM Na2ATP in presence of 4 mM MgCl2. Movement of the microsphere was measured using a differential interferometer (Denk and Webb, 1990; Stelick et al., 1998). Images of microsphere movement were also captured at 1 msec intervals using the CCD kinetics camera.

Interferometer data demonstrated microsphere movement at approximately
9.5-10.5 Hz. Following the assumption that the g
subunit moves in three, 120° steps, the rotation of the g
subunit was approximately 3-4 r.p.s. Measurements of the background,
as well as a microsphere in the absence of ATP displayed no signal
using the interferometer. Similar results were obtained when 1
mm streptavidin-coated magnetic microspheres were attached to
the F1-ATPase.
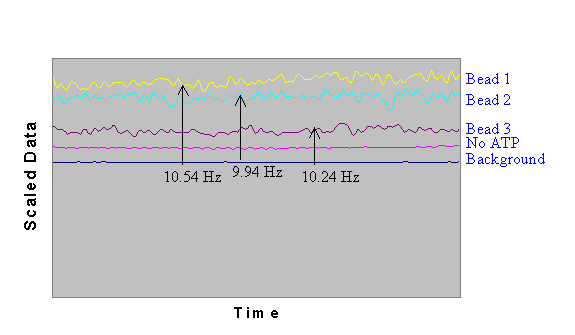
Image analysis demonstrated that microsphere movement occurred
in three discrete steps following a counterclockwise pattern at
a rate of approximately 3-4 r.p.s. Both the interferomenter and
image data confirm a counterclockwise, three step rotational mechanism
of hydrolysis previously reported by Yashuda et al. (1998).

Microsphere movement ceased approximately 40 minutes following
the initial addition of ATP. Prior to stopping, microspheres would
remain at rest for periods up to approximately 600 msec, followed
by 500 msec of continuous movement. We attributed this pattern
of movement to low concentration of ATP in solution. Continuous
movement was reinitiated following the addition of fresh ATP to
the flow cell, suggesting that movement of the microsphere (rotation
of the g subunit) is dependent upon
the presence of ATP.
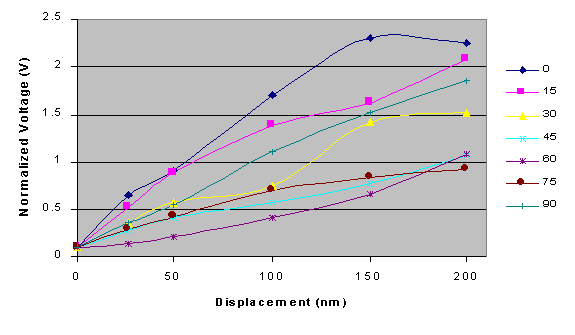
In order to more accurately measure the radial displacement
of a microsphere, calibration of the interferometer was required
to translate changes in voltage into displacement. Calibration
and sensitivity determinations of the differential interferometer
were performed using 1 mm His-tagged fluorescent microspheres
that were attached to nitrocellulose-coated coverslips. As small
as 5 nm movements of the microspheres with a resolution less than
1 nm could be detected by the interferometer. For the expected
range of translation associated with this experiment, the measured
normalized voltage is a linear function of microsphere displacement.

Further examination, however, indicates that changes in voltage
are a function of both the direction and the magnitude of the
displacement. A series of calibration curves were calculated for
microsphere displacement at angles between 0 and 90º, which
clearly illustrate that both distance and angle of displacement
affect the response of the instrument. Therefore, we are currently
adding a quadrant detector that will measure the angular direction
of the microsphere's movement, and permit an accurate quantification
of the real-time deformation of the F1-ATPase
protein during ATP catalysis.
Conclusions
We have established biological and nanofabrication platforms
for the production of organic/inorganic hybrid NEMS. Because of
its size and force generation, F1-ATPase
serves as an excellent model system for evaluating the use of
biomolecular motors in these hybrid devices. The biological platform
that has been established allows us to: (1) easily manipulate
the coding sequence, (2) produce large quantities of protein,
and (3) place specific "handles" on the enzyme for the
precise attachment to nanofabricated devices and substrates. Moreover,
the presented nanofabrication platform permits both the construction
of chemically active sites that are consistent with the size of
the protein and the development of devices that are capable of
translating the energy of a biomolecular motor into useful work.
These results represent the enabling technologies necessary for
integrating NEMS into living organisms.
Further investigation of the engineering properties and motor
performance are necessary for the production of functional nanomechanical
devices powered by F1-ATPase.
For example, the impact of waste products such as heat and protons
on motor performance must be evaluated. Motor performance must
be evaluated as a function of heat, pH, load, and local environmental
conditions. Moreover, dissecting the interaction between the subunits
of the a3b3g complex may allow us to specifically engineer
the protein for increased performance as a biomolecular motor.
These efforts will provide a significant step toward the seamless
integration of nanoscale technologies into living system, and
are central to the creation of organic/inorganic intelligent systems.
References
- Bianchet, M. A., Hullihen, J., Pedersen, P. L., and Amzel,
L. M. (1998) Proc. Natl. Acad. Sci. USA, pages 11065-11070. The
2.8-Å structure of rat liver F1-ATPase: Configuration of
a critical intermediate in ATP synthesis/hydrolysis.
- Carr, D. W., and Craighead, H. G. (1997) J. Vac. Sci. Technol.
B 15, pages 2760-2763. Fabrication of nanoelectromechanical systems
in single crystal silicon using silicon on insulator substrates
and electron beam lithography.
- Craighead, H. G., and Mankiewich, P. M. (1982). J. Appl.
Phys. 53, pages 7186-7188. Ultra-small metal particle arrays
produced by high resolution electron-beam lithography.
- Denk, W., and Webb W. W. (1990). Applied Optics 29, pages
2382-2391. Optical measurement of picometer displacements of
transparent microscopic objects.
- Ekuni, A., Watanabe, H., Kuroda, N., Sawada, K., Murakami,
H., and Kanazawa, H. (1998) FEBS Letters 427, pages 64-68. Reconstitution
of F1-ATPase activity from Escherichia coli subunits a,
b, and subunit g tagged with
histidine residues at the C-terminus.
- Kinosita Jr., K., Yashuda, R., Noji, H., Ishiwata, S., and
Yoshida, M. (1998) Cell 93: 21-24. F1-ATPase: A rotary motor
made of a single molecule.
- Lercel, M. J., Redinbo, G. F., Rooks, F. M., Tiberio, R.
C., and Craighead, H. G. (1995) Microelectron. Eng., pages 43-46.
Electron beam nanofabrication with self-assembled monolayers
of alkylthiols and alkylsiloxanes.
- Lercel, M. J., H. G. Craighead, A. N. Parikh, K. Seshadri
and D. L. Allara (1996). Appl. Phys. Lett. 68, pages 1504-1506.
Sub-10 nm lithography with self-assembled monolayers.
- Matsui, T. and Yoshida, M. (1995) Biochim. Biophys. Acta
1231, pages 139-146. Expression of the wild-type and Cys-/Trp-less
a3b3g complex of the thermophilic F1-ATPase in Escherichia coli.
- Matsui, T., Muneyuki, E., Honda, M., Allison, W., Dou, C.,
and Yoshida, M. (1997) J. Biol. Chem., pages 8215-8221. Catalytic
activity of the a3b3g
complex of F1-ATPase without noncatalytic nucleotide binding
sites.
- Noji, H., Yashuda, R., Yoshida, M., and Kinosita Jr., K.
(1997) Nature 386, pages 299-302. Direct observation of the rotation
of F1-ATPase.
- Sabbert, D., Engelbrecht, S, and Junge, W. (1996) Nature
381, pages 623-625. Intersubunit rotation in active F1-ATPase.
- Stelick, S. J., Montemagno, C. D., and Bachand, G. D. (1998)
Rev. Sci. Instrum. 69: (in press). Detection of single molecule
rotary movement using fluorescence imaging.
- Yashuda, R., Noji, H., Kinosita Jr., K., and Yoshida, M.
(1998) Cell 93, pages 1117-1124. F1-ATPase is a highly efficient
molecular motor that rotates with discrete 120° steps.
|