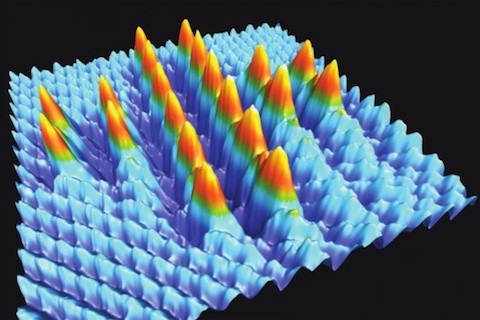
If the above picture reminds you of something like it some 27 years ago when physicists announced a nanostructure built atom-by-atom, then it is important to recognize there are multiple crucial differences between the above 2014 image of a Swiss cross formed from 20 precisely placed bromine atoms and the 1990 image of the IBM… Continue reading Building atom-by-atom on insulator at room temperature
Building atom-by-atom on insulator at room temperature